Peripheral membrane protein
Peripheral membrane proteins, or extrinsic membrane proteins,[1] are membrane proteins that adhere only temporarily to the biological membrane with which they are associated. These proteins attach to integral membrane proteins, or penetrate the peripheral regions of the lipid bilayer. The regulatory protein subunits of many ion channels and transmembrane receptors, for example, may be defined as peripheral membrane proteins. In contrast to integral membrane proteins, peripheral membrane proteins tend to collect in the water-soluble component, or fraction, of all the proteins extracted during a protein purification procedure. Proteins with GPI anchors are an exception to this rule and can have purification properties similar to those of integral membrane proteins.
The reversible attachment of proteins to biological membranes has shown to regulate cell signaling and many other important cellular events, through a variety of mechanisms.[2] For example, the close association between many enzymes and biological membranes may bring them into close proximity with their lipid substrate(s).[3] Membrane binding may also promote rearrangement, dissociation, or conformational changes within many protein structural domains, resulting in an activation of their biological activity.[4][5] Additionally, the positioning of many proteins are localized to either the inner or outer surfaces or leaflets of their resident membrane.[6] This facilitates the assembly of multi-protein complexes by increasing the probability of any appropriate protein–protein interactions.

Binding to the lipid bilayer
[edit]
Peripheral membrane proteins may interact with other proteins or directly with the lipid bilayer. In the latter case, they are then known as amphitropic proteins.[4] Some proteins, such as G-proteins and certain protein kinases, interact with transmembrane proteins and the lipid bilayer simultaneously. Some polypeptide hormones, antimicrobial peptides, and neurotoxins accumulate at the membrane surface prior to locating and interacting with their cell surface receptor targets, which may themselves be peripheral membrane proteins.
The phospholipid bilayer that forms the cell surface membrane consists of a hydrophobic inner core region sandwiched between two regions of hydrophilicity, one at the inner surface and one at the outer surface of the cell membrane (see lipid bilayer article for a more detailed structural description of the cell membrane). The inner and outer surfaces, or interfacial regions, of model phospholipid bilayers have been shown to have a thickness of around 8 to 10 Å, although this may be wider in biological membranes that include large amounts of gangliosides or lipopolysaccharides.[7] The hydrophobic inner core region of typical biological membranes may have a thickness of around 27 to 32 Å, as estimated by Small angle X-ray scattering (SAXS).[8] The boundary region between the hydrophobic inner core and the hydrophilic interfacial regions is very narrow, at around 3 Å, (see lipid bilayer article for a description of its component chemical groups). Moving outwards away from the hydrophobic core region and into the interfacial hydrophilic region, the effective concentration of water rapidly changes across this boundary layer, from nearly zero to a concentration of around 2 M.[9][10] The phosphate groups within phospholipid bilayers are fully hydrated or saturated with water and are situated around 5 Å outside the boundary of the hydrophobic core region.[11]
Some water-soluble proteins associate with lipid bilayers irreversibly and can form transmembrane alpha-helical or beta-barrel channels. Such transformations occur in pore forming toxins such as colicin A, alpha-hemolysin, and others. They may also occur in BcL-2 like protein , in some amphiphilic antimicrobial peptides , and in certain annexins . These proteins are usually described as peripheral as one of their conformational states is water-soluble or only loosely associated with a membrane.[12]
Membrane binding mechanisms
[edit]
The association of a protein with a lipid bilayer may involve significant changes within tertiary structure of a protein. These may include the folding of regions of protein structure that were previously unfolded or a re-arrangement in the folding or a refolding of the membrane-associated part of the proteins. It also may involve the formation or dissociation of protein quaternary structures or oligomeric complexes, and specific binding of ions, ligands, or regulatory lipids.
Typical amphitropic proteins must interact strongly with the lipid bilayer in order to perform their biological functions. These include the enzymatic processing of lipids and other hydrophobic substances, membrane anchoring, and the binding and transfer of small nonpolar compounds between different cellular membranes. These proteins may be anchored to the bilayer as a result of hydrophobic interactions between the bilayer and exposed nonpolar residues at the surface of a protein,[13] by specific non-covalent binding interactions with regulatory lipids , or through their attachment to covalently bound lipid anchors.
It has been shown that the membrane binding affinities of many peripheral proteins depend on the specific lipid composition of the membrane with which they are associated.[14]

Non-specific hydrophobic association
[edit]Amphitropic proteins associate with lipid bilayers via various hydrophobic anchor structures. Such as amphiphilic α-helixes, exposed nonpolar loops, post-translationally acylated or lipidated amino acid residues, or acyl chains of specifically bound regulatory lipids such as phosphatidylinositol phosphates. Hydrophobic interactions have been shown to be important even for highly cationic peptides and proteins, such as the polybasic domain of the MARCKS protein or histactophilin, when their natural hydrophobic anchors are present. [15]
Covalently bound lipid anchors
[edit]Lipid anchored proteins are covalently attached to different fatty acid acyl chains on the cytoplasmic side of the cell membrane via palmitoylation, myristoylation, or prenylation. On the exoplasmic face of the cell membrane, lipid anchored proteins are covalently attached to the lipids glycosylphosphatidylinositol (GPI) and cholesterol.[16][17] Protein association with membranes through the use of acylated residues is a reversible process, as the acyl chain can be buried in a protein's hydrophobic binding pocket after dissociation from the membrane. This process occurs within the beta-subunits of G-proteins. Perhaps because of this additional need for structural flexibility, lipid anchors are usually bound to the highly flexible segments of proteins tertiary structure that are not well resolved by protein crystallographic studies.
Specific protein–lipid binding
[edit]
Some cytosolic proteins are recruited to different cellular membranes by recognizing certain types of lipid found within a given membrane.[18] Binding of a protein to a specific lipid occurs via specific membrane-targeting structural domains that occur within the protein and have specific binding pockets for the lipid head groups of the lipids to which they bind. This is a typical biochemical protein–ligand interaction, and is stabilized by the formation of intermolecular hydrogen bonds, van der Waals interactions, and hydrophobic interactions between the protein and lipid ligand. Such complexes are also stabilized by the formation of ionic bridges between the aspartate or glutamate residues of the protein and lipid phosphates via intervening calcium ions (Ca2+). Such ionic bridges can occur and are stable when ions (such as Ca2+) are already bound to a protein in solution, prior to lipid binding. The formation of ionic bridges is seen in the protein–lipid interaction between both protein C2 type domains and annexins..
Protein–lipid electrostatic interactions
[edit]Any positively charged protein will be attracted to a negatively charged membrane by nonspecific electrostatic interactions. However, not all peripheral peptides and proteins are cationic, and only certain sides of membrane are negatively charged. These include the cytoplasmic side of plasma membranes, the outer leaflet of bacterial outer membranes and mitochondrial membranes. Therefore, electrostatic interactions play an important role in membrane targeting of electron carriers such as cytochrome c, cationic toxins such as charybdotoxin, and specific membrane-targeting domains such as some PH domains, C1 domains, and C2 domains.
Electrostatic interactions are strongly dependent on the ionic strength of the solution. These interactions are relatively weak at the physiological ionic strength (0.14M NaCl): ~3 to 4 kcal/mol for small cationic proteins, such as cytochrome c, charybdotoxin or hisactophilin.[15][19][20]
Spatial position in membrane
[edit]Orientations and penetration depths of many amphitropic proteins and peptides in membranes are studied using site-directed spin labeling,[21] chemical labeling, measurement of membrane binding affinities of protein mutants,[22] fluorescence spectroscopy,[23] solution or solid-state NMR spectroscopy,[24] ATR FTIR spectroscopy,[25] X-ray or neutron diffraction,[26] and computational methods.[27][28][29][30]
Two distinct membrane-association modes of proteins have been identified. Typical water-soluble proteins have no exposed nonpolar residues or any other hydrophobic anchors. Therefore, they remain completely in aqueous solution and do not penetrate into the lipid bilayer, which would be energetically costly. Such proteins interact with bilayers only electrostatically, for example, ribonuclease and poly-lysine interact with membranes in this mode. However, typical amphitropic proteins have various hydrophobic anchors that penetrate the interfacial region and reach the hydrocarbon interior of the membrane. Such proteins "deform" the lipid bilayer, decreasing the temperature of lipid fluid-gel transition.[31] The binding is usually a strongly exothermic reaction.[32] Association of amphiphilic α-helices with membranes occurs similarly.[26][33] Intrinsically unstructured or unfolded peptides with nonpolar residues or lipid anchors can also penetrate the interfacial region of the membrane and reach the hydrocarbon core, especially when such peptides are cationic and interact with negatively charged membranes.[34][35][36]
Categories
[edit]Enzymes
[edit]Peripheral enzymes participate in metabolism of different membrane components, such as lipids (phospholipases and cholesterol oxidases), cell wall oligosaccharides (glycosyltransferase and transglycosidases), or proteins (signal peptidase and palmitoyl protein thioesterases). Lipases can also digest lipids that form micelles or nonpolar droplets in water.
| Class | Function | Physiology | Structure |
|---|---|---|---|
| Alpha/beta hydrolase fold | Catalyzes the hydrolysis of chemical bonds.[37] | Includes bacterial, fungal, gastric and pancreatic lipases, palmitoyl protein thioesterases, cutinase, and cholinesterases | central beta sheet inserted in between two layers of alpha helices[38] |
| Phospholipase A2 (secretory and cytosolic) | Hydrolysis of sn-2 fatty acid bond of phospholipids.[39] | Lipid digestion, membrane disruption, and lipid signaling. | contains catalytic amino acid triad: aspartic acid, serine, and histidine[40] |
| Phospholipase C | Hydrolyzes PIP2, a phosphatidylinositol, into two second messagers, inositol triphosphate and diacylglycerol.[41] | Lipid signaling | core structure composed of a split triosephosphate isomerase (TIM) barrel which has an active site, catalytic residues, and a Ca2+ binding site [42] |
| Cholesterol oxidases | Oxidizes and isomerizes cholesterol to cholest-4-en-3-one.[43] | Depletes cellular membranes of cholesterol, used in bacterial pathogenesis. | two loops of residue which act as a lid on the active site[44] |
| Carotenoid oxygenase | Cleaves carotenoids.[45] | Carotenoids function in both plants and animals as hormones (includes vitamin A in humans), pigments, flavors, floral scents and defense compounds. | composed of multiple enzymes attached together forming branch-like structures[46] |
| Lipoxygenases | Iron-containing enzymes that catalyze the dioxygenation of polyunsaturated fatty acids.[47] | In animals lipoxygenases are involved in the synthesis of inflammatory mediators known as leukotrienes. | hundreds of amino acids that makes up a protein are organized into two domains: beta-sheet N terminal and helical C terminal[48] |
| Alpha toxins | Cleave phospholipids in the cell membrane, similar to Phospholipase C.[49] | Bacterial pathogenesis, particularly by Clostridium perfringens. | soluble monomer with oligomeric pre-pore complexes[50] |
| Sphingomyelinase C | A phosphodiesterase, cleaves phosphodiester bonds.[51] | Processing of lipids such as sphingomyelin. | saposin domain and connector regions with a metallophosphate catalytic domain [52] |
| Glycosyltransferases: MurG and Transglycosidases | Catalyzes the transfer of sugar moieties from activated donor molecules to specific acceptor molecules, forming glycosidic bonds.[53] | Biosynthesis of disaccharides, oligosaccharides and polysaccharides (glycoconjugates), MurG is involved in bacterial peptidoglycan biosynthesis. | three glycine rich loops: one in the C terminal and two in the N terminal [54] |
| Ferrochelatase | Converts protoporphyrin IX into heme.[55] | Involved in porphyrin metabolism, protoporphyrins are used to strengthen egg shells. | polypeptide folded into two domains that each have a four-stranded parallel beta sheet flanked by alpha helices[56] |
| Myotubularin-related protein family | Lipid phosphatase that dephosphorylates PtdIns3P and PtdIns(3,5)P2.[57] | Required for muscle cell differentiation. | contains a GRAM domain, SET interacting domain, and a PDZ binding domain[58] |
| Dihydroorotate dehydrogenases | Oxidation of dihydroorotate (DHO) to orotate.[59] | Biosynthesis of pyrimidine nucleotides in prokaryotic and eukaryotic cells. | composed of two domains: alpha/beta barrel domain that contains the active site and an alpha-helical domain that forms the opening tunnel to the active site [60] |
| Glycolate oxidase | Catalyses the oxidation of α-hydroxycarboxylic acids to the corresponding α-ketoacids.[61] | In green plants, the enzyme participates in photorespiration. In animals, the enzyme participates in production of oxalate. | β8/α8 fold containing alpha helices, beta strands, and loops and turns[62] |
Membrane-targeting domains (“lipid clamps")
[edit]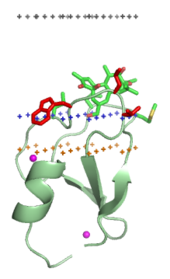
Membrane-targeting domains associate specifically with head groups of their lipid ligands embedded into the membrane. These lipid ligands are present in different concentrations in distinct types of biological membranes (for example, PtdIns3P can be found mostly in membranes of early endosomes, PtdIns(3,5)P2 in late endosomes, and PtdIns4P in the Golgi).[18] Hence, each domain is targeted to a specific membrane.
- C1 domains and phorbol esters.
- C2 domains bind phosphatidylserine, phosphatidylcholine or PtdIns(3,4)P2 or PtdIns(4,5)P2.
- Pleckstrin homology domains, PX domains, and Tubby domains bind different phosphoinositides
- FYVE domains are more specific for PtdIns3P.
- ENTH domains bind PtdIns(3,4)P2 or PtdIns(4,5)P2.
- ANTH domain binds PtdIns(4,5)P2.
- Proteins from ERM (ezrin/radixin/moesin) family bind PtdIns(4,5)P2.
- Other phosphoinositide-binding proteins include phosphotyrosine-binding domain and certain PDZ domains. They bind PtdIns(4,5)P2.
- Discoidin domains of blood coagulation factors
- ENTH, VHS and ANTH domains
Structural domains
[edit]Structural domains mediate attachment of other proteins to membranes. Their binding to membranes can be mediated by calcium ions (Ca2+) that form bridges between the acidic protein residues and phosphate groups of lipids, as in annexins or GLA domains.
| Class | Function | Physiology | Structure |
|---|---|---|---|
| Annexins | Calcium-dependent intracellular membrane/ phospholipid binding.[63] | Functions include vesicle trafficking, membrane fusion and ion channel formation. | |
| Synapsin I | Coats synaptic vesicles and binds to several cytoskeletal elements.[64] | Functions in the regulation of neurotransmitter release. | |
| Synuclein | Unknown cellular function.[65] | Thought to play a role in regulating the stability and/or turnover of the plasma membrane. Associated with both Parkinson's disease and Alzheimer's disease. | |
| GLA-domains of the coagulation system | Gamma-carboxyglutamate (GLA) domains are responsible for the high-affinity binding of calcium ions.[66] | Involved in function of clotting factors in the blood coagulation cascade. | |
| Spectrin and α-actinin-2 | Found in several cytoskeletal and microfilament proteins.[67] | Maintenance of plasma membrane integrity and cytoskeletal structure. |
Transporters of small hydrophobic molecules
[edit]These peripheral proteins function as carriers of non-polar compounds between different types of cell membranes or between membranes and cytosolic protein complexes. The transported substances are phosphatidylinositol, tocopherol, gangliosides, glycolipids, sterol derivatives, retinol, fatty acids, water, macromolecules, red blood cells, phospholipids, and nucleotides.
- Glycolipid transfer proteins
- Lipocalins including retinol binding proteins and fatty acid-binding proteins
- Polyisoprenoid-binding protein, such as YceI protein domain
- Ganglioside GM2 activator proteins
- CRAL-TRIO domain (α-Tocopherol and phosphatidylinositol sec14p transfer proteins)
- Sterol carrier proteins
- Phosphatidylinositol transfer proteins and STAR domains
- Oxysterol-binding protein
Electron carriers
[edit]These proteins are involved in electron transport chains. They include cytochrome c, cupredoxins, high potential iron protein, adrenodoxin reductase, some flavoproteins, and others.
Polypeptide hormones, toxins, and antimicrobial peptides
[edit]Many hormones, toxins, inhibitors, or antimicrobial peptides interact specifically with transmembrane protein complexes. They can also accumulate at the lipid bilayer surface, prior to binding their protein targets. Such polypeptide ligands are often positively charged and interact electrostatically with anionic membranes.
Some water-soluble proteins and peptides can also form transmembrane channels. They usually undergo oligomerization, significant conformational changes, and associate with membranes irreversibly. 3D structure of one such transmembrane channel, α-hemolysin, has been determined. In other cases, the experimental structure represents a water-soluble conformation that interacts with the lipid bilayer peripherally, although some of the channel-forming peptides are rather hydrophobic and therefore were studied by NMR spectroscopy in organic solvents or in the presence of micelles.
| Class | Proteins | Physiology |
|---|---|---|
| Venom toxins |
|
Well known types of biotoxins include neurotoxins, cytotoxins, hemotoxins and necrotoxins. Biotoxins have two primary functions: predation (snake, scorpion and cone snail toxins) and defense (honeybee and ant toxins).[68] |
| Sea anemone toxins |
|
Inhibition of sodium and potassium channels and membrane pore formation are the primary actions of over 40 known Sea anemone peptide toxins. Sea anemone are carnivorous animals and use toxins in predation and defense; anemone toxin is of similar toxicity as the most toxic organophosphate chemical warfare agents.[69] |
| Bacterial toxins |
|
Microbial toxins are the primary virulence factors for a variety of pathogenic bacteria. Some toxins, are Pore forming toxins that lyse cellular membranes. Other toxins inhibit protein synthesis or activate second messenger pathways causing dramatic alterations to signal transduction pathways critical in maintaining a variety of cellular functions. Several bacterial toxins can act directly on the immune system, by acting as superantigens and causing massive T cell proliferation, which overextends the immune system. Botulinum toxin is a neurotoxin that prevents neuro-secretory vesicles from docking/fusing with the nerve synapse plasma membrane, inhibiting neurotransmitter release.[70] |
| Fungal toxins |
|
These peptides are characterized by the presence of an unusual amino acid, α-aminoisobutyric acid, and exhibit antibiotic and antifungal properties due to their membrane channel-forming activities.[71] |
| Antimicrobial peptides |
|
The modes of action by which antimicrobial peptides kill bacteria is varied and includes disrupting membranes, interfering with metabolism, and targeting cytoplasmic components. In contrast to many conventional antibiotics these peptides appear to be bacteriocidal instead of bacteriostatic. |
| Defensins |
|
Defensins are a type of antimicrobial peptide; and are an important component of virtually all innate host defenses against microbial invasion. Defensins penetrate microbial cell membranes by way of electrical attraction, and form a pore in the membrane allowing efflux, which ultimately leads to the lysis of microorganisms.[72] |
| Neuronal peptides |
|
These proteins excite neurons, evoke behavioral responses, are potent vasodilatators, and are responsible for contraction in many types of smooth muscle.[73] |
| Apoptosis regulators | Members of the Bcl-2 family govern mitochondrial outer membrane permeability. Bcl-2 itself suppresses apoptosis in a variety of cell types including lymphocytes and neuronal cells. |
See also
[edit]References
[edit]- ^ "extrinsic protein | biology | Britannica". www.britannica.com. Retrieved 2022-07-04.
- ^ Cafiso DS (2005). "Structure and interactions of C2 domains at membrane surfaces". In Tamm LK (ed.). Protein-Lipid Interactions: From Membrane Domains to Cellular Networks. Chichester: John Wiley & Sons. pp. 403–22. ISBN 3-527-31151-3.
- ^ Ghosh M, Tucker DE, Burchett SA, Leslie CC (November 2006). "Properties of the Group IV phospholipase A2 family". Progress in Lipid Research. 45 (6): 487–510. doi:10.1016/j.plipres.2006.05.003. PMID 16814865.
- ^ a b Johnson JE, Cornell RB (2002). "Amphitropic proteins: regulation by reversible membrane interactions (review)". Molecular Membrane Biology. 16 (3): 217–235. doi:10.1080/096876899294544. PMID 10503244.
- ^ Thuduppathy GR, Craig JW, Kholodenko V, Schon A, Hill RB (June 2006). "Evidence that membrane insertion of the cytosolic domain of Bcl-xL is governed by an electrostatic mechanism". Journal of Molecular Biology. 359 (4): 1045–1058. doi:10.1016/j.jmb.2006.03.052. PMC 1785297. PMID 16650855.
- ^ Takida S, Wedegaertner PB (June 2004). "Exocytic pathway-independent plasma membrane targeting of heterotrimeric G proteins". FEBS Letters. 567 (2–3): 209–213. Bibcode:2004FEBSL.567..209T. doi:10.1016/j.febslet.2004.04.062. PMID 15178324. S2CID 36940600.
- ^ McIntosh TJ, Vidal A, Simon SA (2003). "The energetics of peptide-lipid interactions: modification by interfacial dipoles and cholesterol". Current Topics in Membranes. Vol. 52. Academic Press. pp. 205–253. ISBN 978-0-12-643871-0.
- ^ Mitra K, Ubarretxena-Belandia I, Taguchi T, Warren G, Engelman DM (March 2004). "Modulation of the bilayer thickness of exocytic pathway membranes by membrane proteins rather than cholesterol". Proceedings of the National Academy of Sciences of the United States of America. 101 (12): 4083–4088. Bibcode:2004PNAS..101.4083M. doi:10.1073/pnas.0307332101. PMC 384699. PMID 15016920.
- ^ Marsh D (July 2001). "Polarity and permeation profiles in lipid membranes". Proceedings of the National Academy of Sciences of the United States of America. 98 (14): 7777–7782. Bibcode:2001PNAS...98.7777M. doi:10.1073/pnas.131023798. PMC 35418. PMID 11438731.
- ^ Marsh D (December 2002). "Membrane water-penetration profiles from spin labels". European Biophysics Journal. 31 (7): 559–562. doi:10.1007/s00249-002-0245-z. PMID 12602343. S2CID 36212541.
- ^ Nagle JF, Tristram-Nagle S (November 2000). "Structure of lipid bilayers". Biochimica et Biophysica Acta (BBA) - Reviews on Biomembranes. 1469 (3): 159–195. doi:10.1016/S0304-4157(00)00016-2. PMC 2747654. PMID 11063882.
- ^ Goñi FM (2002). "Non-permanent proteins in membranes: when proteins come as visitors (Review)". Molecular Membrane Biology. 19 (4): 237–245. doi:10.1080/0968768021000035078. PMID 12512770. S2CID 20892603.
- ^ Goforth RL, Chi AK, Greathouse DV, Providence LL, Koeppe RE, Andersen OS (May 2003). "Hydrophobic coupling of lipid bilayer energetics to channel function". The Journal of General Physiology. 121 (5): 477–493. doi:10.1085/jgp.200308797. PMC 2217378. PMID 12719487.
- ^ McIntosh TJ, Simon SA (2006). "Roles of bilayer material properties in function and distribution of membrane proteins". Annual Review of Biophysics and Biomolecular Structure. 35 (1): 177–198. doi:10.1146/annurev.biophys.35.040405.102022. PMID 16689633.
- ^ a b Hanakam F, Gerisch G, Lotz S, Alt T, Seelig A (August 1996). "Binding of hisactophilin I and II to lipid membranes is controlled by a pH-dependent myristoyl-histidine switch". Biochemistry. 35 (34): 11036–11044. doi:10.1021/bi960789j. PMID 8780505.
- ^ Silvius JR (2003). "Lipidated peptides as tools for understanding the membrane interactions of lipid-modified proteins". Current Topics in Membranes. Vol. 52. Academic Press. pp. 371–395. ISBN 978-0-12-643871-0.
- ^ Baumann NA, Mennon AK (2002). "Lipid modifications of proteins". In Vance DE, Vance JE (eds.). Biochemistry of Lipids, Lipoproteins and Membranes (4th ed.). Elsevier Science. pp. 37–54. ISBN 978-0-444-51139-3.
- ^ a b Cho W, Stahelin RV (June 2005). "Membrane-protein interactions in cell signaling and membrane trafficking". Annual Review of Biophysics and Biomolecular Structure. 34: 119–151. doi:10.1146/annurev.biophys.33.110502.133337. PMID 15869386.
- ^ Ben-Tal N, Honig B, Miller C, McLaughlin S (October 1997). "Electrostatic binding of proteins to membranes. Theoretical predictions and experimental results with charybdotoxin and phospholipid vesicles". Biophysical Journal. 73 (4): 1717–1727. Bibcode:1997BpJ....73.1717B. doi:10.1016/S0006-3495(97)78203-1. PMC 1181073. PMID 9336168.
- ^ Sankaram MB, Marsh D (1993). "Protein-lipid interactions with peripheral membrane proteins". In Watts A (ed.). Protein-lipid interactions. Elsevier. pp. 127–162. ISBN 0-444-81575-9.
- ^ Malmberg NJ, Falke JJ (2005). "Use of EPR power saturation to analyze the membrane-docking geometries of peripheral proteins: applications to C2 domains". Annual Review of Biophysics and Biomolecular Structure. 34 (1): 71–90. doi:10.1146/annurev.biophys.34.040204.144534. PMC 3637887. PMID 15869384.
- ^ Spencer AG, Thuresson E, Otto JC, Song I, Smith T, DeWitt DL, et al. (November 1999). "The membrane binding domains of prostaglandin endoperoxide H synthases 1 and 2. Peptide mapping and mutational analysis". The Journal of Biological Chemistry. 274 (46): 32936–32942. doi:10.1074/jbc.274.46.32936. PMID 10551860.
- ^ Lathrop B, Gadd M, Biltonen RL, Rule GS (March 2001). "Changes in Ca2+ affinity upon activation of Agkistrodon piscivorus piscivorus phospholipase A2". Biochemistry. 40 (11): 3264–3272. doi:10.1021/bi001901n. PMID 11258945.
- ^ Kutateladze T, Overduin M (March 2001). "Structural mechanism of endosome docking by the FYVE domain". Science. 291 (5509): 1793–1796. Bibcode:2001Sci...291.1793K. doi:10.1126/science.291.5509.1793. PMID 11230696.
- ^ Tatulian SA, Qin S, Pande AH, He X (September 2005). "Positioning membrane proteins by novel protein engineering and biophysical approaches". Journal of Molecular Biology. 351 (5): 939–947. doi:10.1016/j.jmb.2005.06.080. PMID 16055150.
- ^ a b Hristova K, Wimley WC, Mishra VK, Anantharamiah GM, Segrest JP, White SH (July 1999). "An amphipathic alpha-helix at a membrane interface: a structural study using a novel X-ray diffraction method". Journal of Molecular Biology. 290 (1): 99–117. doi:10.1006/jmbi.1999.2840. PMID 10388560.
- ^ Murray D, Honig B (January 2002). "Electrostatic control of the membrane targeting of C2 domains". Molecular Cell. 9 (1): 145–154. doi:10.1016/S1097-2765(01)00426-9. PMID 11804593.
- ^ Efremov RG, Nolde DE, Konshina AG, Syrtcev NP, Arseniev AS (September 2004). "Peptides and proteins in membranes: what can we learn via computer simulations?". Current Medicinal Chemistry. 11 (18): 2421–2442. doi:10.2174/0929867043364496. PMID 15379706.
- ^ Lomize AL, Pogozheva ID, Lomize MA, Mosberg HI (June 2006). "Positioning of proteins in membranes: a computational approach". Protein Science. 15 (6): 1318–1333. doi:10.1110/ps.062126106. PMC 2242528. PMID 16731967.
- ^ Lomize A, Lomize M, Pogozheva I. "Comparison with experimental data". Orientations of Proteins in Membranes. University of Michigan. Retrieved 2007-02-08.
- ^ Papahadjopoulos D, Moscarello M, Eylar EH, Isac T (September 1975). "Effects of proteins on thermotropic phase transitions of phospholipid membranes". Biochimica et Biophysica Acta (BBA) - Biomembranes. 401 (3): 317–335. doi:10.1016/0005-2736(75)90233-3. PMID 52374.
- ^ Seelig J (November 2004). "Thermodynamics of lipid-peptide interactions". Biochimica et Biophysica Acta (BBA) - Biomembranes. 1666 (1–2): 40–50. doi:10.1016/j.bbamem.2004.08.004. PMID 15519307.
- ^ Darkes MJ, Davies SM, Bradshaw JP (1997). "Interaction of tachykinins with phospholipid membranes: A neutron diffraction study". Physica B. 241: 1144–1147. Bibcode:1997PhyB..241.1144D. doi:10.1016/S0921-4526(97)00811-9.
- ^ Ellena JF, Moulthrop J, Wu J, Rauch M, Jaysinghne S, Castle JD, Cafiso DS (November 2004). "Membrane position of a basic aromatic peptide that sequesters phosphatidylinositol 4,5 bisphosphate determined by site-directed spin labeling and high-resolution NMR". Biophysical Journal. 87 (5): 3221–3233. Bibcode:2004BpJ....87.3221E. doi:10.1529/biophysj.104.046748. PMC 1304792. PMID 15315949.
- ^ Marcotte I, Dufourc EJ, Ouellet M, Auger M (July 2003). "Interaction of the neuropeptide met-enkephalin with zwitterionic and negatively charged bicelles as viewed by 31P and 2H solid-state NMR". Biophysical Journal. 85 (1): 328–339. Bibcode:2003BpJ....85..328M. doi:10.1016/S0006-3495(03)74477-4. PMC 1303088. PMID 12829487.
- ^ Zhang W, Crocker E, McLaughlin S, Smith SO (June 2003). "Binding of peptides with basic and aromatic residues to bilayer membranes: phenylalanine in the myristoylated alanine-rich C kinase substrate effector domain penetrates into the hydrophobic core of the bilayer". The Journal of Biological Chemistry. 278 (24): 21459–21466. doi:10.1074/jbc.M301652200. PMID 12670959.
- ^ "Pfam entry Abhydrolase 1". Archived from the original on 2007-09-29. Retrieved 2007-01-25.
- ^ Bauer, Tabea L.; Buchholz, Patrick C. F.; Pleiss, Jürgen (March 2020). "The modular structure of α/β-hydrolases". The FEBS Journal. 287 (5): 1035–1053. doi:10.1111/febs.15071. ISSN 1742-464X. PMID 31545554.
- ^ "Pfam entry: Phospholipase A2". Archived from the original on 2007-09-29. Retrieved 2007-01-25.
- ^ Casale, Jarett; Kacimi, Salah Eddine O.; Varacallo, Matthew (2023), "Biochemistry, Phospholipase A2", StatPearls, Treasure Island (FL): StatPearls Publishing, PMID 30521272, retrieved 2023-11-29
- ^ "Pfam entry: Phosphatidylinositol-specific phospholipase C, X domain". Archived from the original on 2007-09-29. Retrieved 2007-01-25.
- ^ "Phospholipase C", Wikipedia, 2023-08-16, retrieved 2023-11-29
- ^ "Pfam entry: Cholesterol oxidase". Archived from the original on 2007-09-29. Retrieved 2007-01-25.
- ^ Yue, Q. Kimberley; Kass, Ignatius J.; Sampson, Nicole S.; Vrielink, Alice (1999-04-01). "Crystal Structure Determination of Cholesterol Oxidase from Streptomyces and Structural Characterization of Key Active Site Mutants". Biochemistry. 38 (14): 4277–4286. doi:10.1021/bi982497j. ISSN 0006-2960. PMID 10194345.
- ^ "Pfam entry: Retinal pigment epithelial membrane protein". Archived from the original on 2007-09-29. Retrieved 2007-01-25.
- ^ "Carotenoid oxygenase", Wikipedia, 2023-11-29, retrieved 2023-11-29
- ^ "Pfam entry: Lipoxygenase". Archived from the original on 2007-09-29. Retrieved 2007-01-25.
- ^ Prigge, S. T.; Boyington, J. C.; Faig, M.; Doctor, K. S.; Gaffney, B. J.; Amzel, L. M. (1997-11-01). "Structure and mechanism of lipoxygenases". Biochimie. 79 (11): 629–636. doi:10.1016/S0300-9084(97)83495-5. ISSN 0300-9084. PMID 9479444.
- ^ PDBsum entry: Alpha Toxin
- ^ "Alpha Toxin - an overview | ScienceDirect Topics". www.sciencedirect.com. Retrieved 2023-11-29.
- ^ "Pfam entry: Type I phosphodiesterase". Archived from the original on 2007-09-29. Retrieved 2007-01-25.
- ^ Xiong, Zi-Jian; Huang, Jingjing; Poda, Gennady; Pomès, Régis; Privé, Gilbert G. (2016-07-31). "Structure of Human Acid Sphingomyelinase Reveals the Role of the Saposin Domain in Activating Substrate Hydrolysis". Journal of Molecular Biology. 428 (15): 3026–3042. doi:10.1016/j.jmb.2016.06.012. ISSN 0022-2836. PMID 27349982.
- ^ "Pfam entry: Glycosyl transferases group 1". Archived from the original on 2007-09-29. Retrieved 2007-01-25.
- ^ Ünligil, Uluğ M; Rini, James M (2000-10-01). "Glycosyltransferase structure and mechanism". Current Opinion in Structural Biology. 10 (5): 510–517. doi:10.1016/S0959-440X(00)00124-X. ISSN 0959-440X. PMID 11042447.
- ^ "Pfam entry: Ferrochelatase". Archived from the original on 2007-09-29. Retrieved 2007-01-25.
- ^ Al-Karadaghi, Salam; Hansson, Mats; Nikonov, Stanislav; Jönsson, Bodil; Hederstedt, Lars (November 1997). "Crystal structure of ferrochelatase: the terminal enzyme in heme biosynthesis". Structure. 5 (11): 1501–1510. doi:10.1016/s0969-2126(97)00299-2. ISSN 0969-2126. PMID 9384565.
- ^ "Pfam entry:Myotubularin-related". Archived from the original on 2007-09-26. Retrieved 2007-01-25.
- ^ Emery and Rimoin's Principles and Practice of Medical Genetics. ISBN 978-0-12-383834-6. Retrieved 2023-11-29.
- ^ "Pfam entry:Dihydroorotate dehydrogenase". Archived from the original on 2007-09-26. Retrieved 2007-01-25.
- ^ Liu, Shenping; Neidhardt, Edie A; Grossman, Trudy H; Ocain, Tim; Clardy, Jon (January 2000). "Structures of human dihydroorotate dehydrogenase in complex with antiproliferative agents". Structure. 8 (1): 25–33. doi:10.1016/s0969-2126(00)00077-0. ISSN 0969-2126. PMID 10673429.
- ^ "Pfam entry: FMN-dependent dehydrogenase". Archived from the original on 2007-09-29. Retrieved 2007-01-25.
- ^ "Glycolate oxidase - Proteopedia, life in 3D". proteopedia.org. Retrieved 2023-11-28.
- ^ "Pfam entry: Annexin". Archived from the original on 2007-09-29. Retrieved 2007-01-25.
- ^ "Pfam entry Synapsin N". Archived from the original on 2007-09-26. Retrieved 2007-01-25.
- ^ "Pfam entry Synuclein". Archived from the original on 2007-09-26. Retrieved 2007-01-25.
- ^ "Pfam entry: Gla". Archived from the original on 2007-09-29. Retrieved 2007-01-25.
- ^ "Pfam entry Spectrin". Archived from the original on 2007-09-26. Retrieved 2007-01-25.
- ^ Rochat H, Martin-Eauclaire MF, eds. (2000). Animal toxins: facts and protocols. Basel: Birkhũser Verlag. ISBN 3-7643-6020-8.
- ^ Patocka J, Strunecka A (1999). "Sea Anemone Toxins". The ASA Newsletter. Archived from the original on 15 June 2013.
- ^ Schmitt CK, Meysick KC, O'Brien AD (1999). "Bacterial toxins: friends or foes?". Emerging Infectious Diseases. 5 (2): 224–234. doi:10.3201/eid0502.990206. PMC 2640701. PMID 10221874.
- ^ Chugh JK, Wallace BA (August 2001). "Peptaibols: models for ion channels". Biochemical Society Transactions. 29 (Pt 4): 565–570. doi:10.1042/BST0290565. PMID 11498029.
- ^ Oppenheim JJ, Biragyn A, Kwak LW, Yang D (November 2003). "Roles of antimicrobial peptides such as defensins in innate and adaptive immunity". Annals of the Rheumatic Diseases. 62 (Suppl 2): ii17 – ii21. doi:10.1136/ard.62.suppl_2.ii17. PMC 1766745. PMID 14532141.
- ^ "Pfam entry Tachykinin". Archived from the original on 2007-09-26. Retrieved 2007-01-25.
Further reading
[edit]- Tamm LK, ed. (2005). Protein-Lipid Interactions: From Membrane Domains to Cellular Networks. Chichester: John Wiley & Sons. ISBN 3-527-31151-3.
- Cho W, Stahelin RV (June 2005). "Membrane-protein interactions in cell signaling and membrane trafficking". Annual Review of Biophysics and Biomolecular Structure. 34 (1): 119–151. doi:10.1146/annurev.biophys.33.110502.133337. PMID 15869386.
- Goñi FM (2002). "Non-permanent proteins in membranes: when proteins come as visitors (Review)". Molecular Membrane Biology. 19 (4): 237–245. doi:10.1080/0968768021000035078. PMID 12512770. S2CID 20892603.
- Johnson JE, Cornell RB (1999). "Amphitropic proteins: regulation by reversible membrane interactions (review)". Molecular Membrane Biology. 16 (3): 217–235. doi:10.1080/096876899294544. PMID 10503244.
- Seaton BA, Roberts MF (1996). "Peripheral membrane proteins". In Mertz K, Roux B (eds.). Biological Membranes. Boston, MA: Birkhauser. pp. 355–403.
- Benga G (1985). "Protein-lipid interactions in biological membranes". In Benga G (ed.). Structure and Properties of Biological Membranes. Vol. 1. Boca Raton, FL: CRC Press. pp. 159–188.
- Kessel A, Ben-Tal N (January 2002). "Free energy determinants of peptide association with lipid bilayers". Current Topics in Membranes. 52: 205–253. doi:10.1016/S1063-5823(02)52010-X. ISBN 9780121533526.
- Malmberg NJ, Falke JJ (2005). "Use of EPR power saturation to analyze the membrane-docking geometries of peripheral proteins: applications to C2 domains". Annual Review of Biophysics and Biomolecular Structure. 34 (1): 71–90. doi:10.1146/annurev.biophys.34.040204.144534. PMC 3637887. PMID 15869384.
- McIntosh TJ, Simon SA (2006). "Roles of bilayer material properties in function and distribution of membrane proteins". Annual Review of Biophysics and Biomolecular Structure. 35 (1): 177–198. doi:10.1146/annurev.biophys.35.040405.102022. PMID 16689633.
External links
[edit]- Peripheral membrane proteins in OPM database
- DOLOP Genomics-oriented database of bacterial lipoproteins
- Peptaibol database
- Antimicrobial Peptide Database Archived 2011-07-20 at the Wayback Machine
